Batch Processing
After you found the optimal parameters on an image in the last step, you can press the Run Batch-button to apply the spot-detection and counting to a series of images.
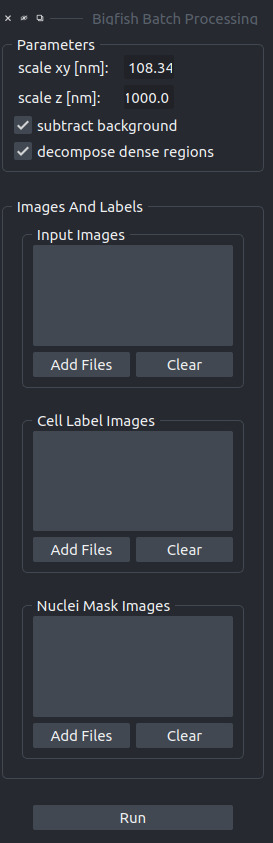
We need to provide the scale, since bigfish does not read ot from the image files.
- scale xy
The scale (voxel size) in the xy-plane in nm.
- scale z
The scale (voxel size) in the z-dimension in nm.
- subtract background
If selected the pre-processing step of background subtraction will be applied to the images.
- decompose dense regions
If selected the decomposition of dense regions will be applied on the result of the initial spot detection.
- Input Images
The images containing the FISH-spots
- Cell Label Images
The images containing the cell labels, in the same order as the input images.
- Nuclei Mask Images
The images containing the nuclei labels or masks, in the same order as the input images and cell label images.
The correspondence between the spot image, the cell label image and the nuclei mask image is made by the position in the list, i.e. the third image in the cell label images-list correponds to the third input image, etc.
Add files to the lists. You can use the Clear-button to empty a list and the context menu to selectively remove images. The Input Images must be provided, the two other lists are optional. If no cell labels are provided, the spots in the whole image will be counted and reported as belonging to the background label 0. If cell labels are provided but no nuclei masks, the spots will be counted by cell, and all spots will be reported to be in the cytoplasm.
Press the Run-button to start the batch-processing. The processing will run in a separate thread and report the progress with a progress bar.