Quick start: A user guide
1. Introduction
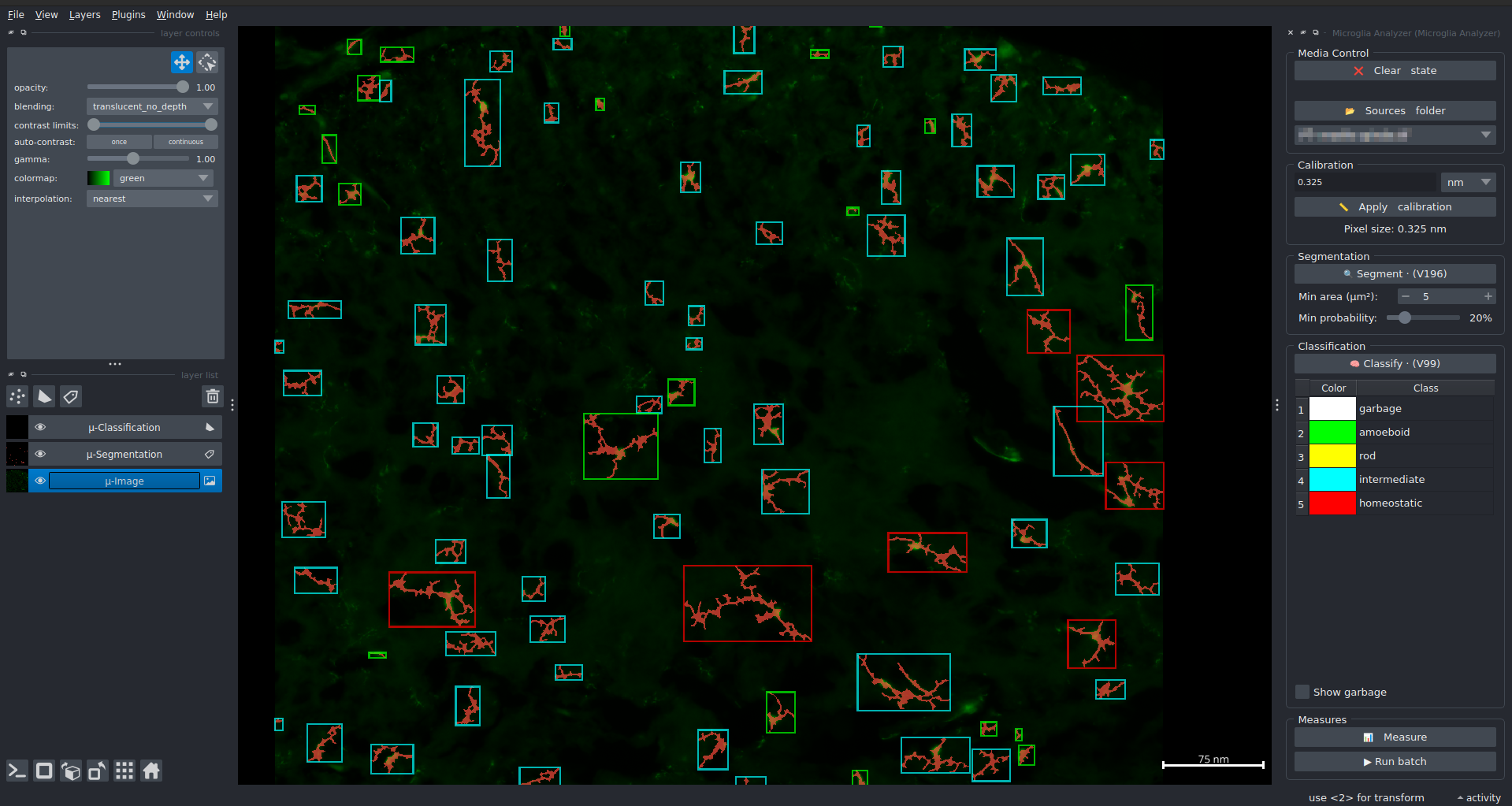
“Microglia Analyzer” is a Python module allowing to segment, classify and measure microglia from 2D fluo images.
This module is also a Napari plugin in case you would need a graphical interface.
The segmentation produced doesn’t allow to recover the actual surface of microglia since the output of the segmentation is a shape optimized for skeletonization.
Each cell is classified according 3 classes: “amoeboid”, “intermediate” and “homeostatic”.
- The measures (bundled in a CSV file by the end) include:
The total length of each cell
The number of junctions for each cell
The number of end points for each cell
The average branch length for each cell
From the GUI (Napari), a batch mode is available allowing you to run the whole workflow on an entire folder.
2. Install the plugin
We strongly recommend to use conda or any other virtual environment manager instead of installing Napari and microglia-analyzer in your system’s Python.
Napari is only required if you want to use microglia-analyzer with a graphical interface.
Napari is not part of microglia-analyzer’s dependencies, so you will have to install it separately.
Each of the commands below is supposed to be run after you activated your virtual environment.
If the install was successful, you should see the plugin in Napari in the top bar menu: Plugins > Microglia Analyzer > Microglia Analyzer.
A. Development versions
Method |
Instructions |
|---|---|
pip |
|
GitHub |
|
From an archive |
|
B. Stable versions
Method |
Instructions |
|---|---|
pip |
Activate your conda environment, and type |
NapariHub |
Go in the plugins menu of Napari and search for “Microglia Analyzer” |
3. Notes
The plugin provides detailed output, so it’s recommended to monitor the terminal if you want detailed information about its actions.
If a crash occurs, please create an issue and include the relevant image(s) and a copy of your terminal for further investigation.
Napari currently supports only open file formats, so make sure to convert your images to TIFF format before using them with Napari.