How to use the “Microglia Analyzer” widget?
1. Get your data ready
From this point, we will only consider that you are using the Napari plugin. If you are using the Python module, you can look at the “__main__” section of the “ma_worker.py” file. This Napari plugin expects your data to respect a precise format:
The images must be available as dissociated TIFF images (no jpg, no 3D stack, no video, …).
The files must have exactly 1 channel. We recommend to keep your images in 16-bits grayscale.
All the images that you want to process at the same time must be in the same folder.
Avoid using special characters in the folder’s name. (Tips: Doranum)
.
├── 📁 my-awesome-experiment
│ ├── some-image.tif
│ ├── another-image.tif
│ ├── ...
│ └── last-image.tif
2. Workflow
a. Import your experiment (Media control)
In this section, you will import your experiment so you can visualize and analyze it.
The
Clear statebutton is useless unless you just analyzed something else and need to reset the plugin.Use the
Sources folderbutton to provide the path to the folder containing your images. It is the path of “my-awesome-experiment” in the previous example.In the drop-down menu below, all the images of your folder should show up. Clicking on any of them in the list should display it in the main viewer.
You can adjust the contrast and brightness in the upper-left corner of Napari.
b. Calibration (Calibration)
Providing measures in physical units requires the plugin to know the size of a pixel.
Just fill the number field with the size of a pixel and select the unit in the drop-down menu next to it before clicking the
Apply calibrationbutton.If your image doesn’t appear in the viewer anymore, or if it becomes too small, you can click on the little “Home” button in the lower-left corner of Napari to center the view.
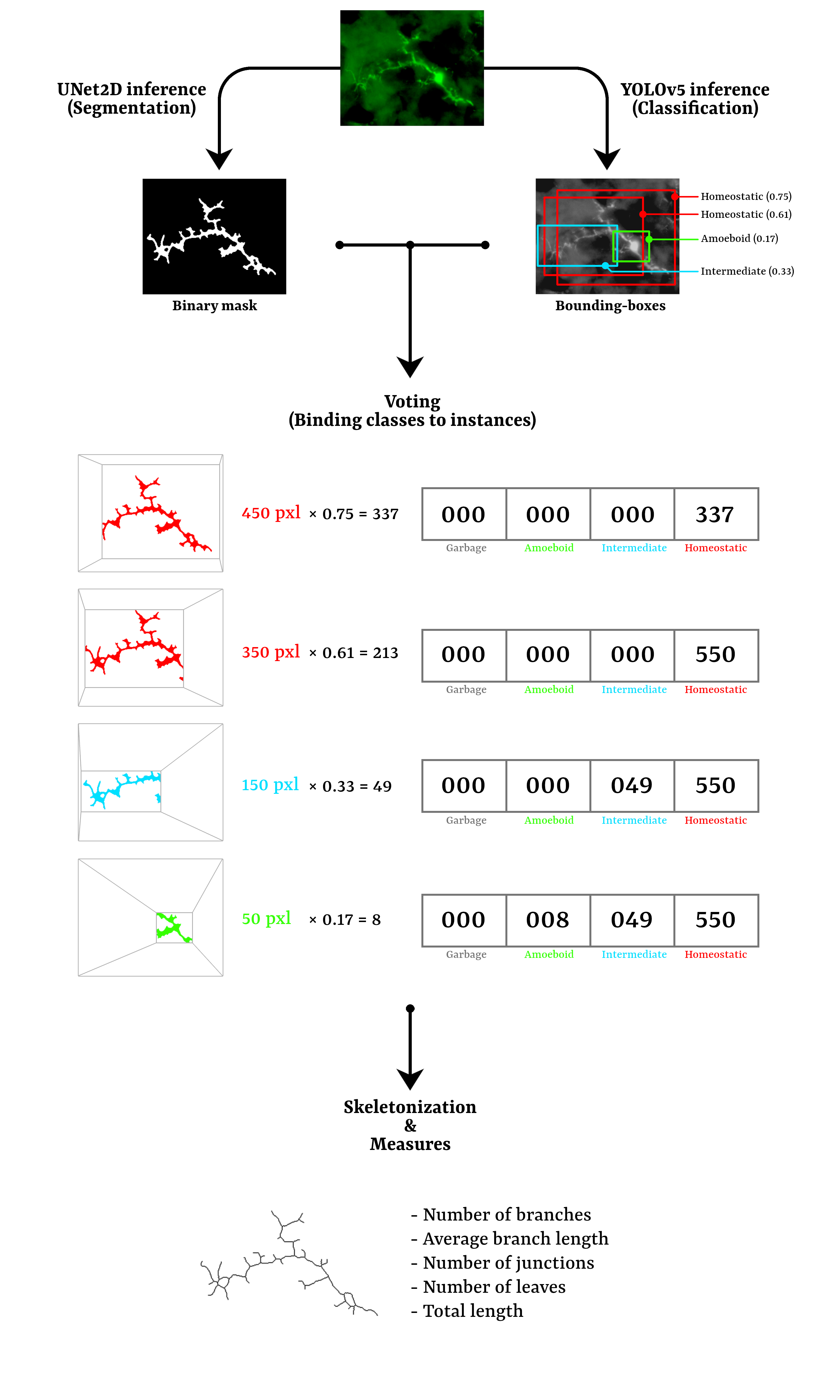
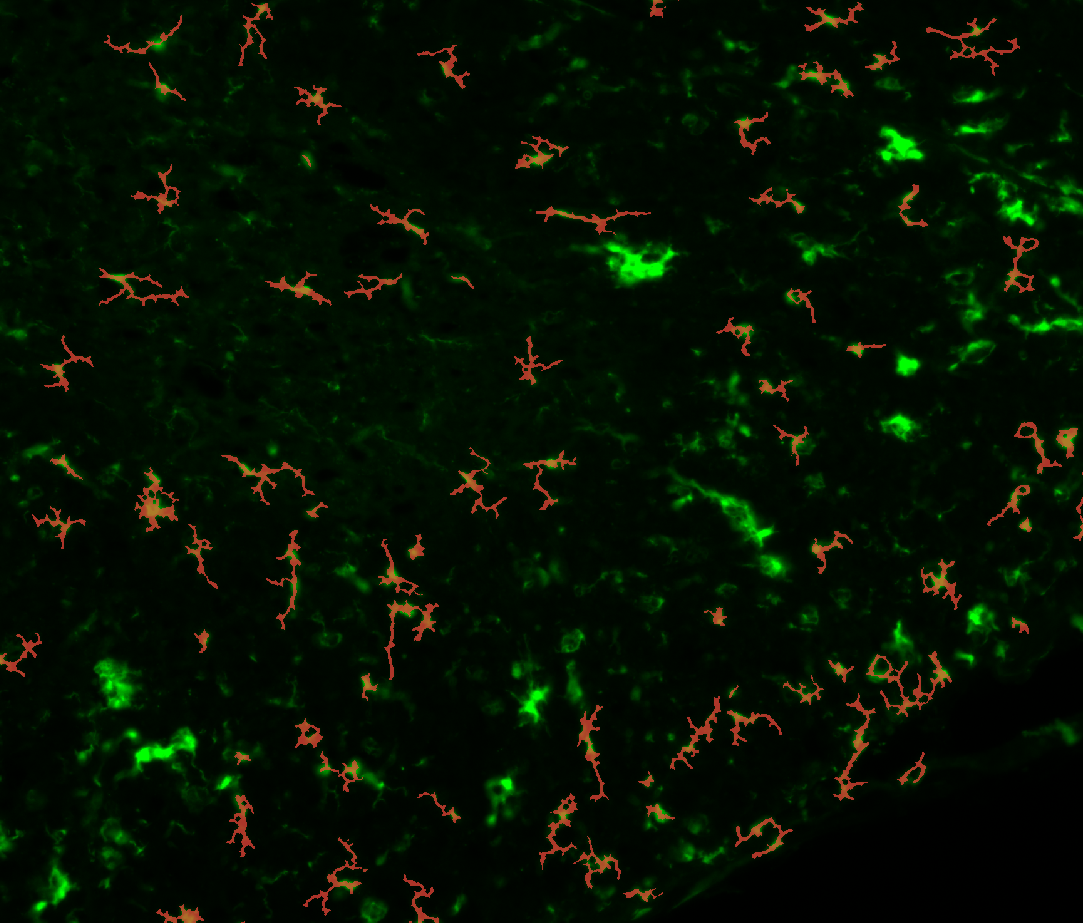
c. Segment the microglia (Segmentation)
Setting |
Description |
|---|---|
Min area (µm²) |
Minimal area that an object must reach to avoid being discarded. |
Min probability |
The raw output of the segmentation process is a probability map that must be thresholded to create a mask. |
You can set the “Min area” and the “Min probability” before or after the segmentation.
You can just click the
Segment microgliabutton to start the segmentation.The first time, the model will have to be downloaded from internet.
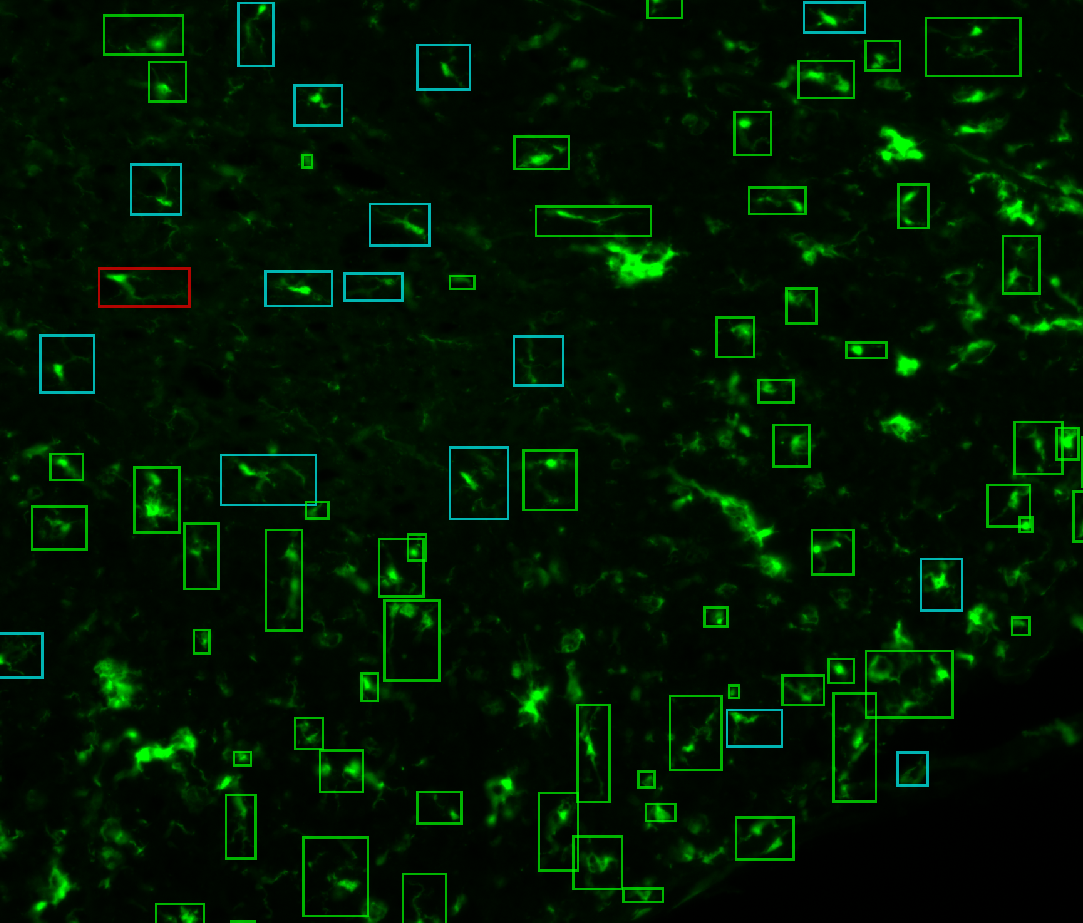
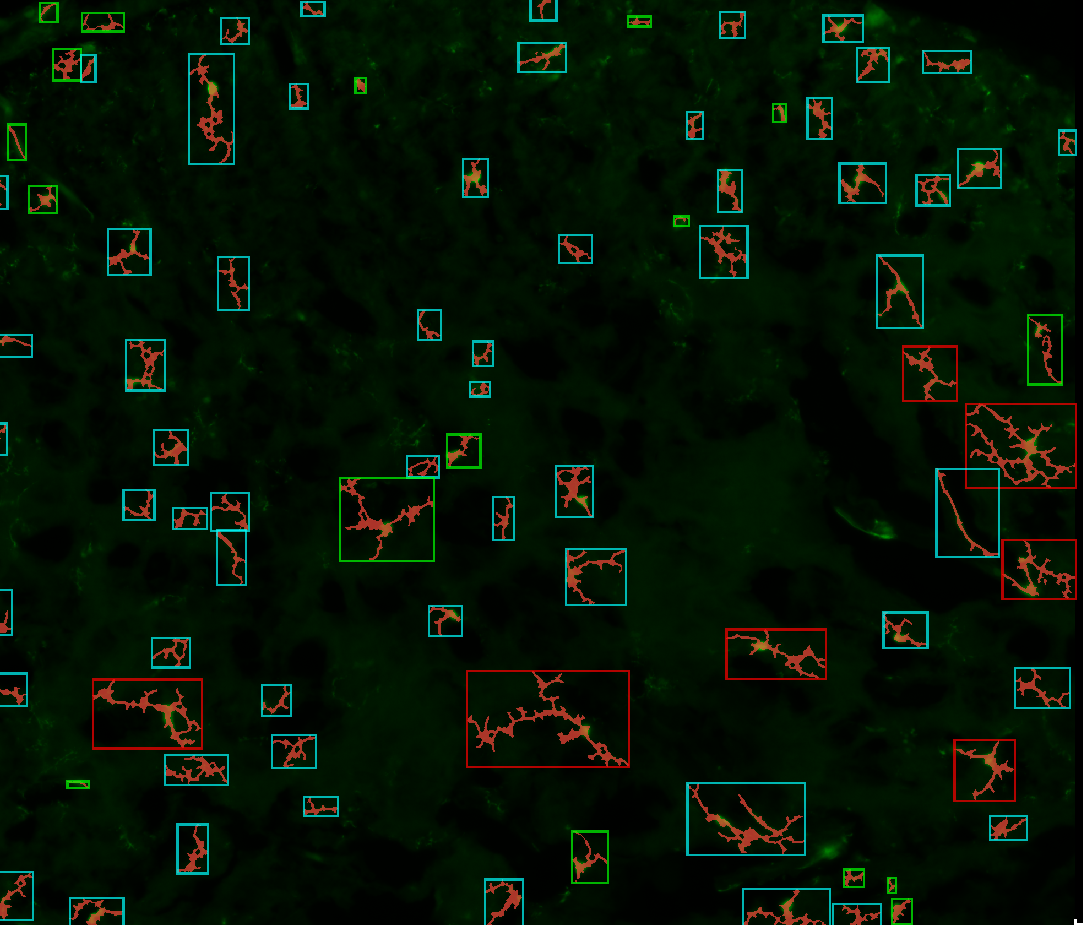
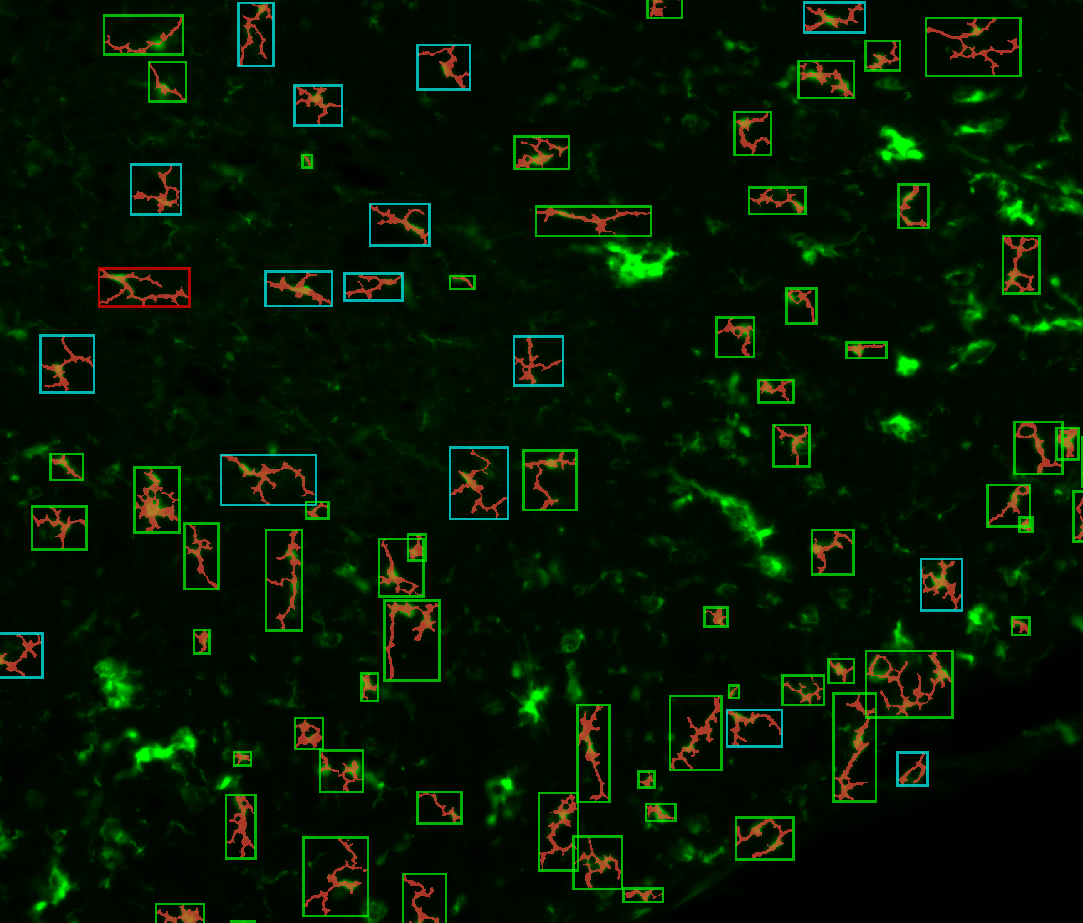
d. Classify the microglia (Classification)
Click the classify button.
The model has to be downloaded from the internet the first time you use it.
By the end of the process, the possible classes will show up below the
Classifybutton, with their associated color.Some elements are classified as “Garbage” (aggregated objects, out-of-focus, filaments from other slices, …) so you can use the
Show garbagecheckbox to hide them.At this point, you must have each microglia surrounded by a colored box, representing its class.
e. Extract measures + batch (Measures)
If you click the
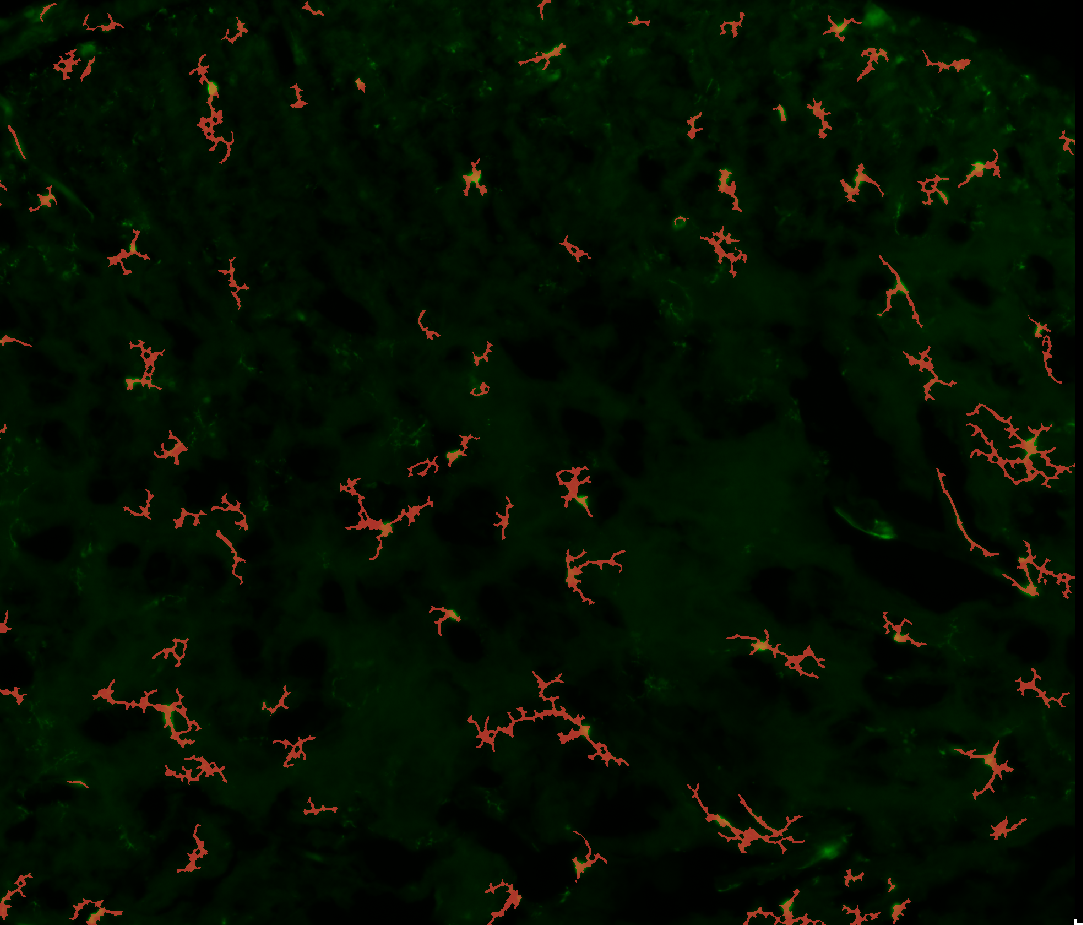
Measurebutton, the plugin will compute the skeleton of each non-garbage object and extract the measures mentioned in the introduction.A new folder named “controls” should appear in the folder containing your images. It should contain a CSV file named after your image as well as a control image.
If everything looks fine, you can click the
Run batchbutton to run the whole workflow over the entire folder using the same settings you just used.The number of images left to process should be displayed on the button now named
Stop batch.By the end of the process, your “controls” folder should contain a control image for each input image, and a unique CSV file (“results.csv”) aggregating the measures of all the images.
3. Examples of processed data
4. Workflow diagram