Quick start: A user guide
1. Install the plugin
Method |
Instructions |
|---|---|
With pip |
Launch your conda environment, and type |
From Napari Hub |
Go in the plugins menu of Napari and search for “Spots in yeasts” |
From GitHub |
Launching your conda environment and then use the command |
2. Quick demonstration
3. Before starting
Spots in yeasts expects a very precise file as its input:
The file must be a TIFF (no .czi, .nd, …)
Both channels must be packed in this TIFF.
The first channel must contain the spots and the second the brightfield.
- The number of slices is arbitrary.
If you have a single slice, it is the one that is going to be used.
If you have a stack, a MIP will be used for the processing. It will be based on the most in-focus slice as well as N slices before and after it.
If your images don’t respect the previous conditions, you can check the page explaining how to conform your data.
4. Settings
If you modify some settings, don’t forget to click the ‘Apply settings’ button for them to take effect!!!
Name |
Description |
|---|---|
LoG Radius |
Radius (in pixels) of the Gaussian filter applied to the spots channel before detection. |
Slices around focus |
Number of slices taken around the focus slice (in the case of a stack). |
Death threshold |
Intensity threshold in the spots channel beyond which a cell is considered dead. |
Min spots distance |
Minimum distance (in pixels) required between two spots (to account for noise). |
Spot area min |
Minimum area of a spot; anything lower than that will be considered as waste. |
Spot area max |
Maximum area of a spot; anything beyond that will be considered as waste. |
Extent threshold |
Minimal extent tolerated before discarding a spot. |
Solidity threshold |
Minimum solidity tolerated before discarding a spot. |
N/C cover threshold |
Percentage of a cell that must be covered by a nucleus for it to be considered dead. |
Intensity threshold |
Percentage of the max intensity in the spots channel required to validate a spot. |
Export format |
Format used to create the exported CSV file. |
5. Processing
a. Process a single image
If something is already present in the Napari viewer, click on
Clear layers. It will flush everything present in your current viewer.Drag’N’drop your TIFF file into the Napari viewer. It should appear in the left column.
Click on
Split channels. Now, your original image should have disappeared (in the left column) and two (or three) new layers took its place.They represent the brightfield, spots’ fluo channel and the optional nuclei’s channel. Layers are opaque, which means that as long as you don’t toggle the little eye icon in the left column, the viewer shows you the uppermost layer.You can now click the
Segment cellsbutton. This operation can take quite a while depending on your hardware. The presence of a dedicated GPU on your machine would help a lot. The result will spawn as a new layer containing a label for each cell in the brightfield. At this point, cells cut by the borders have already been discarded. If you make this new layer active (just click on it in the left column until its name turns blue), you can pass your mouse over each label to verify its value. These values match the indices present in the results file.If your nuclei are marked, you can click the
Segment nucleibutton. It will both segment the nuclei and attempt to merge the mothers with their daughter.Click the
Segment spotsbutton. This step is much quicker. You can zoom on your image to see where spots were detected. Note that the white circles only represent the position of detected spots, they do not represent the segmentation. If you prefer to see the segmentation, you can hide thespots-positionslayer and use thelabeled-spotsone instead. Of the same way as in the previous step, you can make this layer active to see the indices, which are the same as in the results file. If your nuclei are stained, the color of the markers depends on the category of the spot (cytoplasmic, peripheral, nuclear).Finally, you can use the
Extract statsbutton. This will open a CSV file in your default spreadsheet software. Keep in mind that it is a temporary file, it is not saved anywhere and will disappear as soon as you close it if you don’t save it.
b. Batch processing
Start by making a folder in which you place all the images that you need to get processed. Note that a unique CSV file is produced for the whole batch, so make sure that your folder contains images that can be compared to each other (same acquisition conditions), otherwise, you may end up with inconsistant data.
Now, you need to create the folder that will receive the control images produced by the plugin. Indeed, in case you observe odd results, the plugin produces a bunch of control images in batch mode that you can inspect if required. It is not required that the output folder is empty, but it is recommended though.
Then, you can set your input and output path.
Finally, you can click the
Run batchbutton.In Napari’s GUI, you can click on
activityin the lower right corner to see a progress bar. You can monitor way more precisely what is going on thanks to the terminal that tells in real time what is being done.At the end of the execution, the produced CSV is located at the root of the output folder. Each folder ending with
.yscis a set of control images for one input image.
c. Results control
Each control is a folder (instead of an image)
You can simply drag’n’drop the folder (not its content) into Napari instead of opening each image that it contains. A reader recognizing folders ending with
.ysc(yeasts spots control) is bundled with the plugin.In there, each cell is represented by its outline.
d. How to read the results?
source: The name of the image (without its extension) from which the following data was extracted.cell-index: A unique number assigned to each cell. This number corresponds to the label on the control image. Please note that some numbers may be missing from the list if cells were cut by the image’s border (resulting in their labels being erased).spot-index: Similarly to ‘cell-index’, this value corresponds to the label on the control image.area: The number of pixels covered by the spot.intensity-mean: The average intensity recorded for all the pixels within the spot.intensity-min: The lowest intensity recorded among all the pixels within the spot.intensity-max: The highest intensity recorded among all the pixels within the spot.intensity-sum: Also known as integrated intensity, this is the sum of intensities across all pixels within the spot.perimeter: The length of the perimeter around the spot, measured in steps with diagonal connectivity.solidity: The ratio of the spot’s area to the area of its convex hull. A star-shaped spot will have a value close to 0, while a more circular spot will have a value close to 1.extent: The ratio of the spot’s area to the area of its bounding box, which is the smallest rectangle that contains the spot. This metric gives an idea of how elongated the spot is. For example, a perfect circle and a perfect ellipse will both have a solidity of 1.0, however, their extent will vary.# spots: The number of spots detected in the given cell.
source: The name of the image (without its extension) from which the following data was extracted.cell-index: A unique number assigned to each cell. This number corresponds to the label on the control image. Please note that some numbers may be missing from the list if cells were cut by the image’s border (resulting in their labels being erased).# cytoplasmic spots: Number of spots found exclusively in the cytoplasms.# nuclear spots: Number of spots found exclusively in the nuclei.# peripheral spots: Number of spots found overlaping with both the cytoplasms and the nuclei.
6. Notes
The plugin provides verbose output, so it’s recommended to monitor the terminal if you want detailed information about its actions.
If a crash occurs, please create an issue and include the relevant image(s) for further investigation.
Napari currently supports only open file formats, so make sure to convert your images to TIFF format before using them with Napari.
7. Solidity & Extent
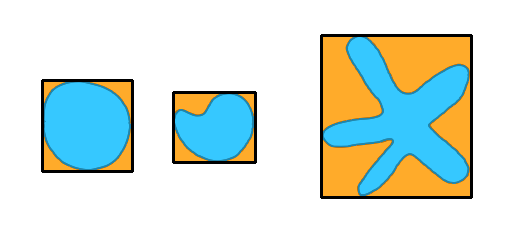 |
The extent is measured as the ratio of the spot's area over its bounding box area. On this illustration, it is represented by the blue area divided by the orange area. |
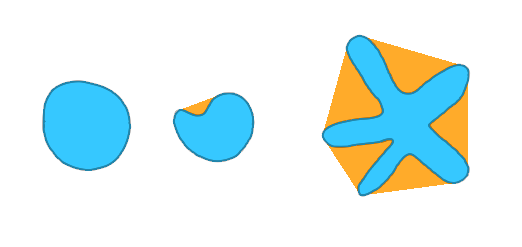 |
The solidity is measured as the ratio of the spot's area over its convex hull area. If the spot is convex (like in the first scenario), the ratio is 1.0. |