Zero Cost Deep-Learning for Microscopy
Volker Bäcker
Montpellier Ressources Imagerie
29/04/2020
Content
- What is ZeroCostDL4Mic?
- Experience Report Noise2Void
- Conclusions
1.1 Spoiler
- Is it really for free?
- yes
- some restrictions, but usable
1.2 What problem does it solve?
- We’ve seen a lot of publications applying deep-learning for microscopy
- But difficult to use for most, because they need to be trained on our data
- need cloud/GPU/cluster access
- need to get the images to the cloud/GPU/cluster
- need programming skills
- need DL basics to select the training parameters
1.3 How does it help?
- prepared jupyter notebooks
- start them on google colab by clicking on a button
- the text walks you through the process step by step
- you don’t have to touch any code
- everything is done via a graphical user interface in the notebook
- access to your data via your google-drive
1.4 What can I do with it?
- There are currently 5 models:
| UNet |
segmentation, EM, brightfield images |
| Stardist (2D/3D) |
nuclei segmentation, DAPI, Hoechst |
| Noise2Void (2D/3D) |
denoising, unsupervised - no ground truth images needed |
| CARE (2D/3D) |
image restoration, low SNR to high SNR |
| fnet |
create fluorescent image from brightfield image |
1.5 How to get started?
- Start at the project’s wiki
- You need
- a google account
- enough free space in your google-drive
- Click on the “Open in colab”-button to start a notebook in colab
- Follow the instructions in the notebook
1.6 What are the restrictions?
- One training/prediction unit can max. take 12h
- colab disconnects every 12 hours
- to make crypto-currency mining impossible
- free google-drive provides 15GB of disk-space
- colab distributes GPUs/TPUs according to availability
- you get access to GPUs/TPUs according to
- availability
- your previous usage
2.1 Noise2Void Experience
- the images (800x800 8-bit)
![]()
2.2 Image Upload
![]()
- upload to google-drive
- 2 images for training, 3 images for predictions
2.3 Running the noise2void
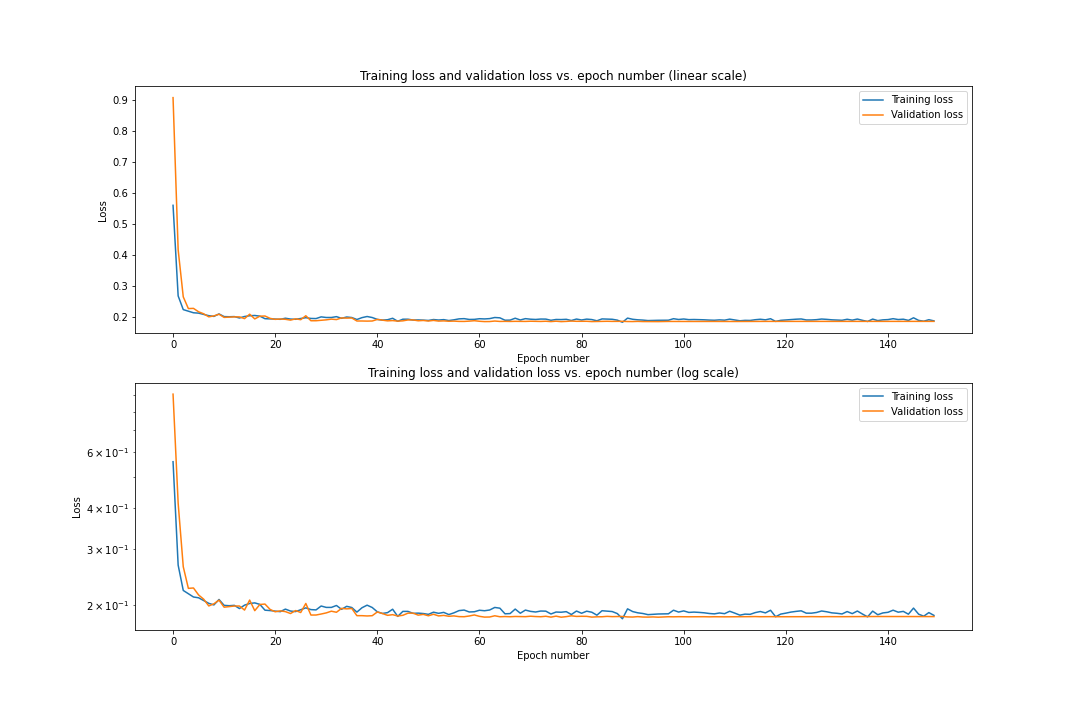
2.4 Training Loss
![]()
2.5 Quality control
- You need ground-truth images to do the quality estimation
- In this case the images are synthetic
- low noise images created with MRI_Create_Synthetic_Spots_Tool
- noise added separately for foreground and background with ImageJ
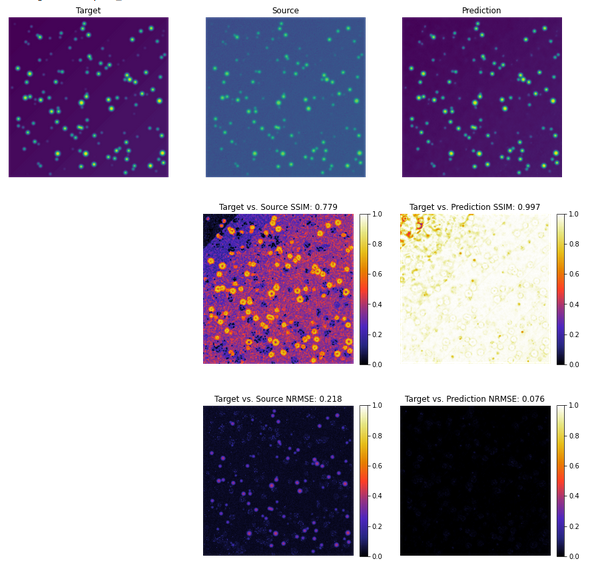
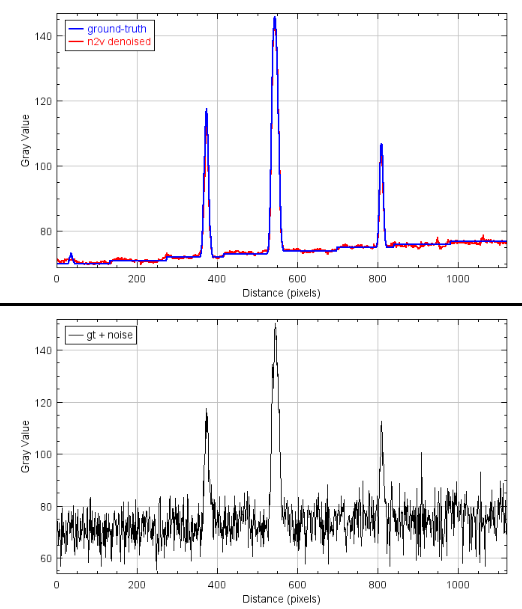
2.6 Quality control results
![]()
- SSIM: structural similarity in [0,1], the closer to one the better
- NRMSE: normalized root mean square error, the closer to 0 the better
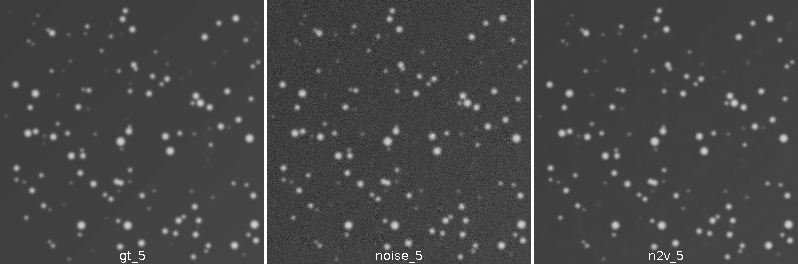
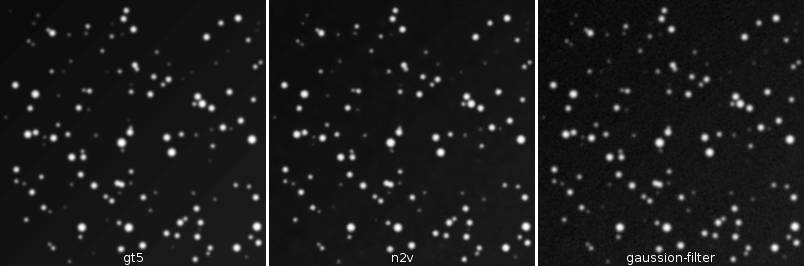
2.7 Results - Images
![]()
![]()
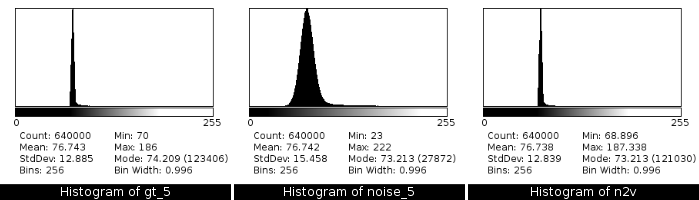
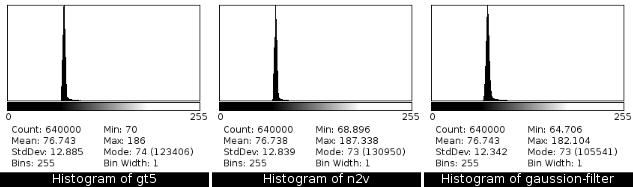
2.8 Results - Histograms
![]()
![]()
2.9 Results - Plots
![]()
2.10 Results - Metrics
![]()
Calculated with the SNR-Plugin [1]
| SNR |
Signal to Noise Ratio |
| PSNR |
Peak Signal to Noise Ratio |
| RMSE |
Root Mean Square Error |
| MAE |
Mean Absolute Error |
[1] D. Sage, M. Unser, Teaching Image-Processing Programming in Java, IEEE Signal Processing Magazine, vol. 20, no. 6, pp. 43-52, November 2003.
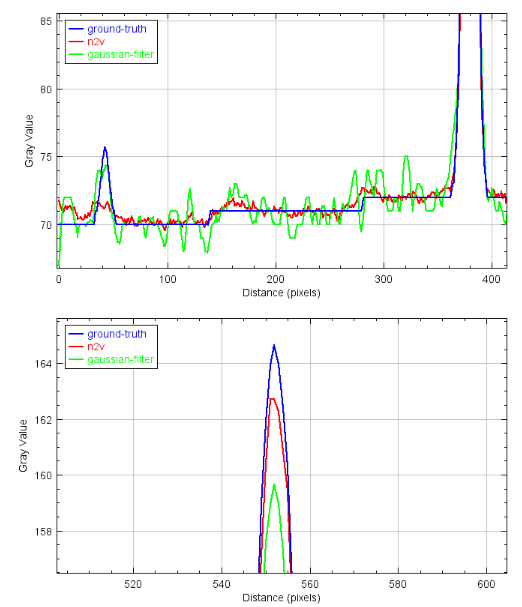
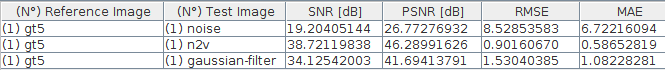
2.11 Comparison with Gaussian-Filter
- Is the n2v denoising better than a simple solution?
2.12 Comparison - Histograms
![]()
![]()
2.13 Comparison - Plots
![]()
2.14 Comparison - Metrics
![]()
Calculated with the SNR-Plugin [1]
| SNR |
Signal to Noise Ratio |
| PSNR |
Peak Signal to Noise Ratio |
| RMSE |
Root Mean Square Error |
| MAE |
Mean Absolute Error |
[1] D. Sage, M. Unser, Teaching Image-Processing Programming in Java, IEEE Signal Processing Magazine, vol. 20, no. 6, pp. 43-52, November 2003.
3.1 Conclusions noise2void experiment
- Training was fast
- The result “looks” very good
- Higher SNR than the Gaussian-blur filter
- Low intensity spots are lost with n2v
- For segmentation Gaussian-blur filter might be better (in this case)
3.2 Conclusions
- We can use Zero Cost Deep-Learning for Microscopy (all)
- We can setup other DeepLearning-models in the same way (the ML-team)
- We can train and apply the model
- on colab
- on analysis pcs (with or without gpus, depending on the model)
- on our own server (we do not have it yet)
- possibly on the meso@lr cluster (not setup yet)
- We can apply the trained models from ImageJ
=> Try it on colab to get started
3.3 An alternative
- ImJoy [1]
- nice environment for deploying DL-models
- also a number of DL-models for microscopy already available
- for cloud computing it uses mybinder (only demo)
- but could user other services
- more complex for setup and data-access
- user friendly gui for training and predictions
[1] Ouyang, W., Mueller, F., Hjelmare, M. et al. ImJoy: an open-source computational platform for the deep learning era. Nat Methods 16, 1199–1200 (2019)