The NapariJ-plugin
Combining the python 3D viewer with FIJI
10.07.2021
Montpellier Ressources Imagerie
Volker Bäcker
Contents
- the 3D spot colocalization example
- what is napari
- napari plugins
- the imagej-python bridge
- the naparij-plugin
- what’s next?
3D spot colocalization
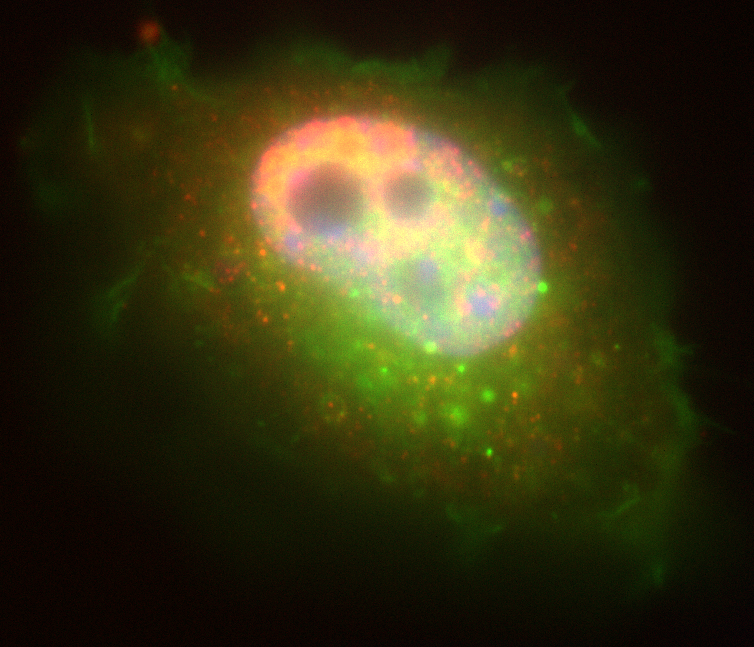
- input:
- 2 spot channels +
nuclei channel
- 2 spot channels +
- objectif:
- for spots not within
the nucleus, calculate
the shortest distances
between spots in
channel one and
channel two
- for spots not within
Solution with Imaris
- Create spots channel one:
- auto-detect + manual selection of ‘quality’ threshold
- Create spots channel two:
- auto-detect + manual selection of ‘quality’ threshold
- Create surface to segment the nucleus
- From the two spot objects creatre binary masks
- Use the nucleus-surface to remove spots within the nucleus in the two mask channels
- Reconvert the spot mask channels to spots
- Find all pairs of spots between the two channels which are closer then a given distance
:) solves the problem :( can not be automatized
Solution with FIJI
- use FeatureJ Laplacian (3D) to create a 3D image in which the minima are the centers of the spots
- use
3D Maxima Finderfrom the3D Suite ImageJ - segment the nucleus via thresholding
- remove points for which the nucleus mask is 255
- remove points for which the quality is below a threshold
- calculate the shortest distance from each point in one channel to the points in the other channel
:) can be automatized :( hard to set a threshold with out seeing the consequences in 3D
Napari
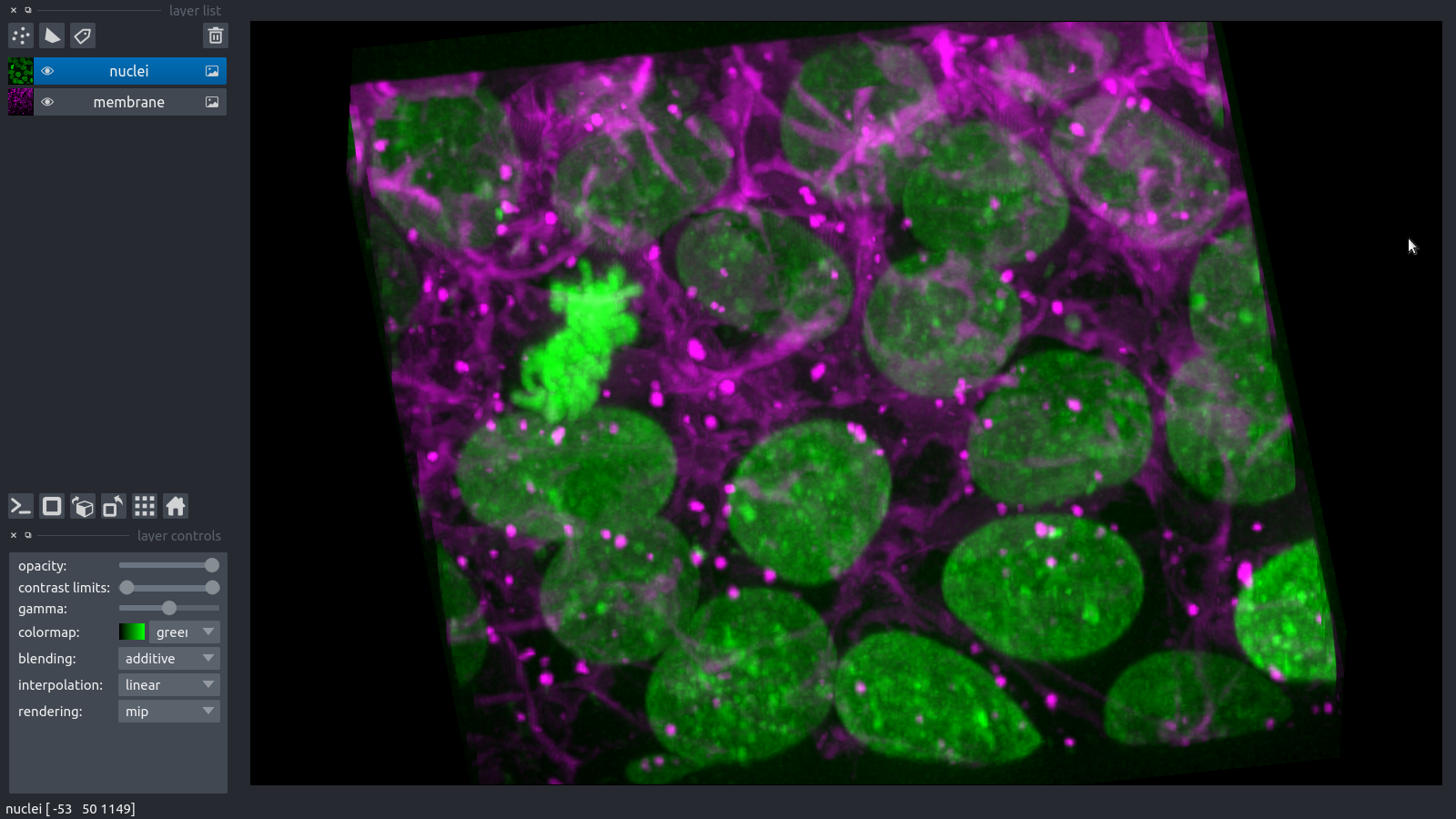
- 3D viewer
- dimensions z, t , … (sliders)
- image layers c
- point, labels, shape layers with gui tools
- surface, track, vector layers via python code
- plugins (widgets or hooks (for example load/save))
- data is an array like python object
- can use dask arrays and zarr-files
- read big data / read remote data
- can use dask arrays and zarr-files
Napari plugins
- plugin can be registered
- registered plugins can be installed from within napari
Napari animation plugin
The ImageJ-python bridge
- Connect ImageJ and python both ways
- Use jpype to start the jvm
- call java from python
- shared memory
- no copying of data
- Use the jupyter-client
- send commands from IJ to the ipython kernel
- the one that started the jvm
- send commands from IJ to the ipython kernel
Example python accessing IJ
data = getResulsTable()
display(data)
cal = IJ.getImage().getCalibration()
coords = np.delete(data.values,[3], axis=1)
icoords = [[z, y, x] for [x,y,z] in coords]
zFactor = cal.getZ(1) / cal.getX(1)
points_layer = viewer.add_points(icoords, size=1, scale=[zFactor, 1, 1])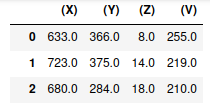
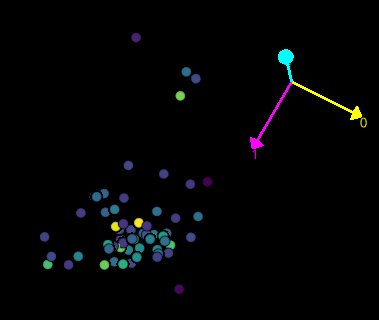
Example IJ accessing python
from ijpb.fiji.IPythonProxy import IPythonProxy
p = IPythonProxy()
p.run("import napari")
p.run("from PyQt5 import QtCore")
p.run("viewer = napari.Viewer()")
p.disconnect()
The naparij-plugin
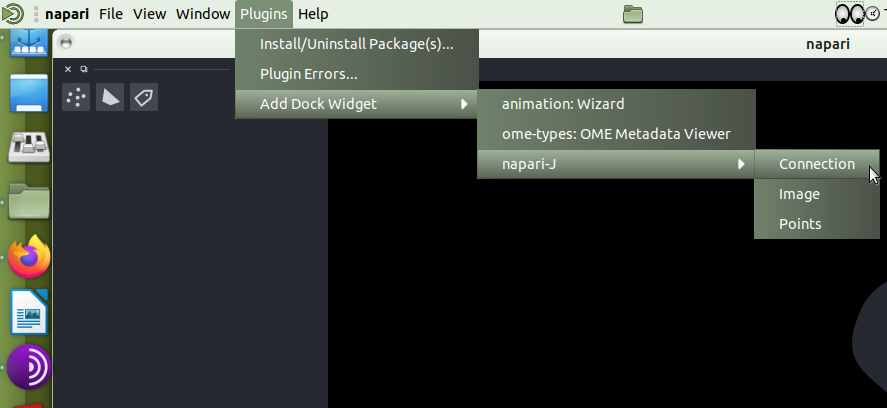
Connection
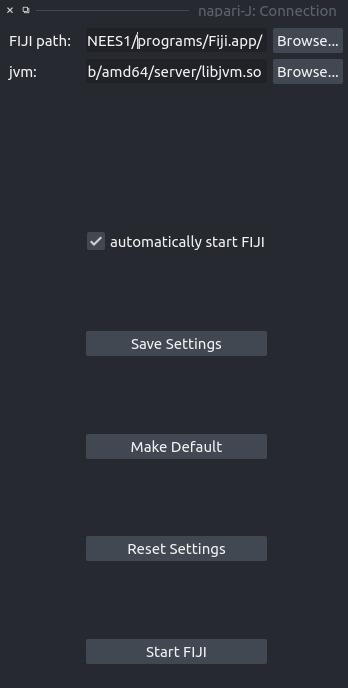
- start the jvm
- set
jupter_connection_file
andpython_executablein IJ- so that the IPython proxy can connect back
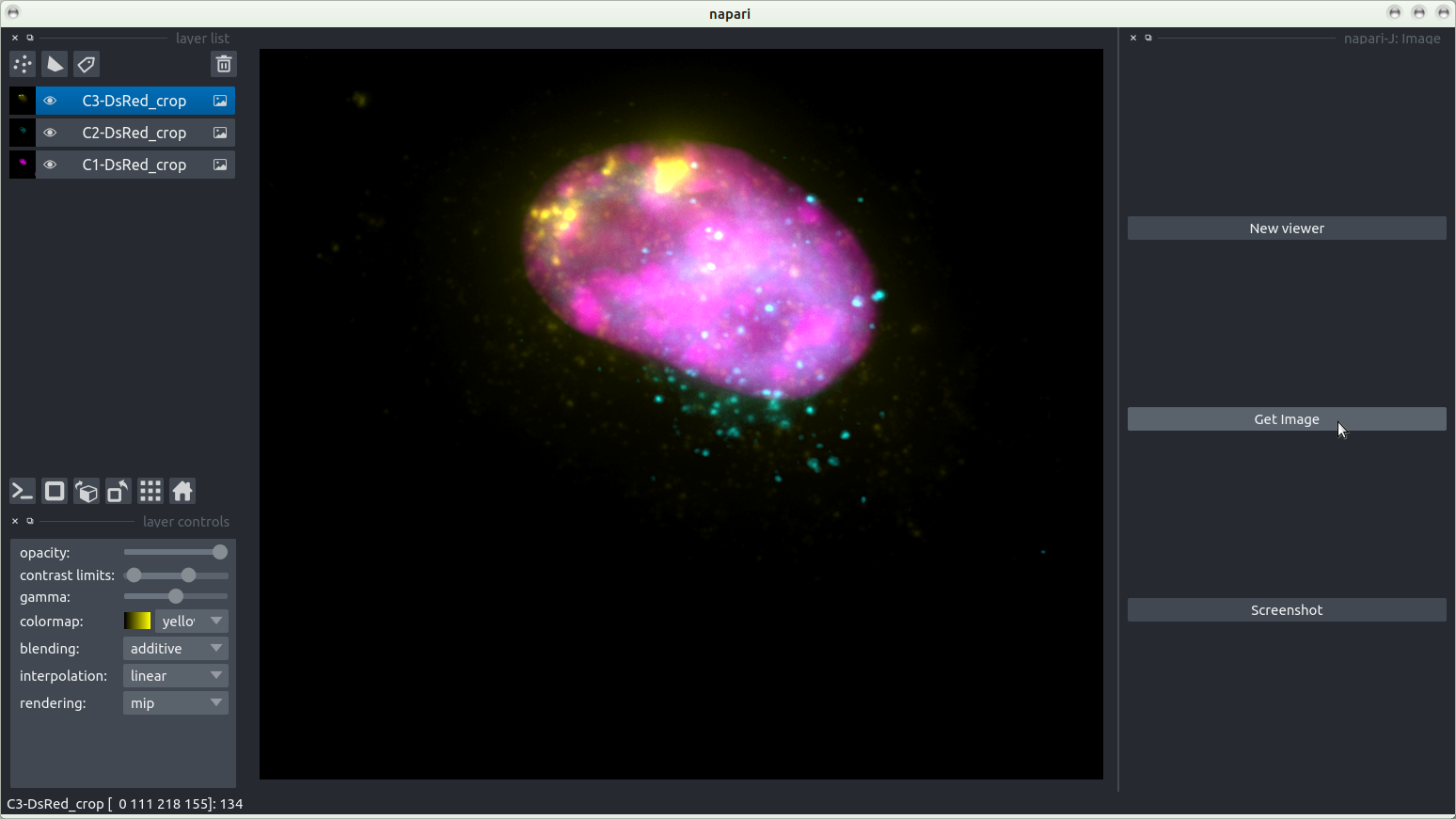
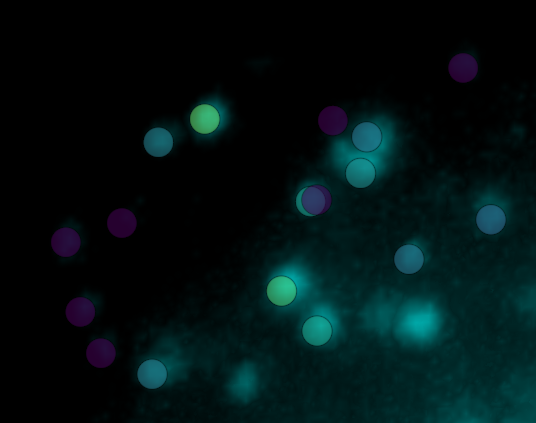
Image
Points
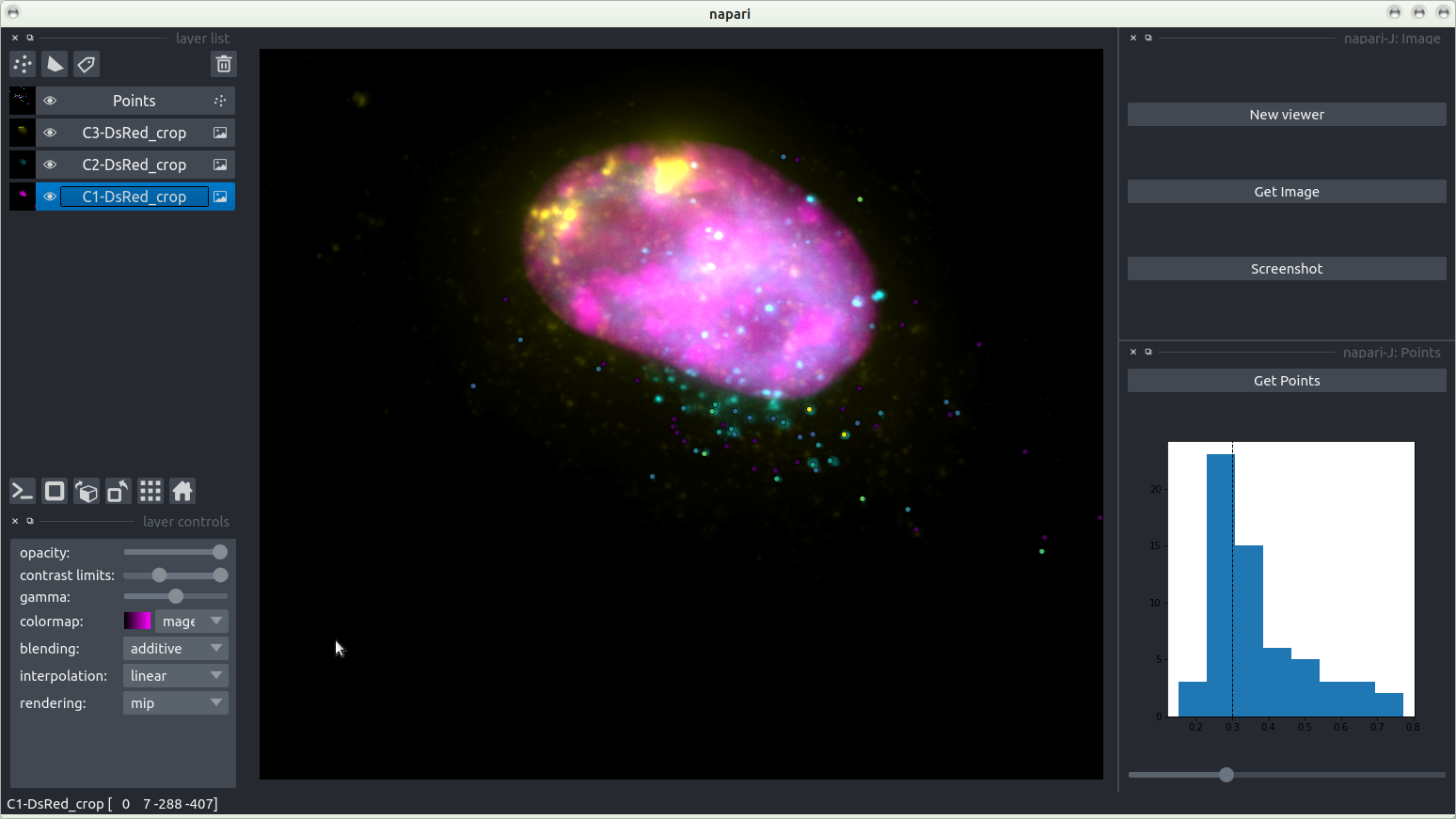
Points (zoom)
What’s next?
- finish the naparij-plugin
- send the filtered points back to IJ
- finish the analysis
- calculate the distances
- Add features to the plugin
- 3D object counter (segmentation) to napari surfaces and back
- tracks, vector-fields, ???, …
- Use the ImageJ-python bridge with other python software
- clustering / ML / DL
- give DL4Mic more control over the pythob process
- IJPB can also be used to connect to a distant IPython via ssh-tunneling
- run on server, in the cloud or on a cluster